Human CALML5 / CLSP Protein (His & GST Tag)
CLSP
- 100ug (NPP3628) Please inquiry
| Catalog Number | P11783-H20E |
|---|---|
| Organism Species | Human |
| Host | E. coli |
| Synonyms | CLSP |
| Molecular Weight | The recombinant human CALML5/GST chimera consists of 388 amino acids and has a calculated molecular mass of 44.2 kDa. It migrates as an approximately 43 kDa band in SDS-PAGE under reducing conditions. |
| predicted N | Met |
| SDS-PAGE | 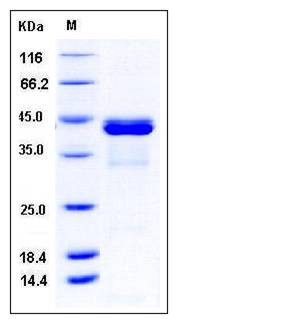 |
| Purity | > 92 % as determined by SDS-PAGE |
| Protein Construction | A DNA sequence encoding the human CALML5 (AAH39172.1) (Met 1-Glu 146) was fused with the N-terminal polyhistidine-tagged GST tag at the N-terminus. |
| Bio-activity | |
| Research Area | Cancer |Signal transduction |Signaling Pathway |Calcium Signaling |Calmodulin Pathway |
| Formulation | Lyophilized from sterile 20mM Tris, 150mM NaCl, 1mM DTT, 0.5mM GSH, 10% glycerol, pH 7.8 1. Normally 5 % - 8 % trehalose, mannitol and 0.01% Tween80 are added as protectants before lyophilization. Specific concentrations are included in the hardcopy of COA. |
| Background | Calmodulin-like protein 5, also known as Calmodulin-like skin protein, CALML5 and CLSP, is a protein which contains four EF-hand domains. CALML5 / CLSP is particularly abundant in the epidermis where its expression is directly related to keratinocyte differentiation.The expression is very low in lung. CALML5 / CLSP binds calcium. It may be involved in terminal differentiation of keratinocytes. Coxsackievirus and adenovirus receptor (CAR) is a member of the immunoglobulin (Ig) superfamily and a component of epithelial tight junction. CAR functions as a primary receptor for coxsackievirus B and adenovirus (Ad) infection. CALML5 / CLSP is closely related to CAR. The structure and dynamics of human calmodulin-like skin protein CALML5 / CLSP have been characterized by NMR spectroscopy. The mobility of CALML5 / CLSP has been found to be different for the N-terminal and C-terminal domains. The N-terminal domain is characterized by four stable helices, which experience large fluctuations. This is shown to be due to mutations in the hydrophobic core. The overall N-terminal domain behavior is similar both in the full-length protein and in the isolated domain. |
| Reference |
