Human CHL-1 Protein (His Tag)
CALL,L1CAM2
- 100ug (NPP2012) Please inquiry
| Catalog Number | P10143-H08H |
|---|---|
| Organism Species | Human |
| Host | Human Cells |
| Synonyms | CALL,L1CAM2 |
| Molecular Weight | The recombinant human CHL1 consists of 1067 amino acids after removal of the signal peptide and predicts a molecular mass of 120 kDa. As a result of glycosylation, the apparent molecular mass of rhCHL1 is approximately 160-180 kDa in SDS-PAGE under non-reduced conditions. |
| predicted N | Ile 25 |
| SDS-PAGE | 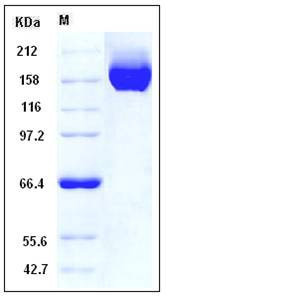 |
| Purity | > 95 % as determined by SDS-PAGE |
| Protein Construction | A DNA sequence encoding the extracellular domain of human CHL1 (AAI04919.1) (Met 1-Gln 1080) was fused with a polyhistidine tag at the C-terminus. |
| Bio-activity | Measured by the ability of the immobilized protein to support the adhesion of C6 Rat brain glial cells. When 5 x 10E4 cells/well are added to CHL1 coated plates (0.8 μg/ml and 100 μl/well), approximately 40%-60% will adhere specifically after 60 minutes at 37℃. |
| Research Area | Neuroscience |Cell Adhesion Proteins |Membrane Proteins |
| Formulation | Lyophilized from sterile PBS, pH 7.4 1. Normally 5 % - 8 % trehalose, mannitol and 0.01% Tween80 are added as protectants before lyophilization. Specific concentrations are included in the hardcopy of COA. |
| Background | Neural cell adhesion molecule L1-like protein, also known as close homolog of L1 (CHL1) is the prototypic member of the CTF / NF-1 family of transcription factors that serve as a novel calcium signaling pathway-responsive transcription factor and is considered as a member of the largest ctf complementation group, consisting of 30 of 126 ctf mutants isolated. CHL1 is a cell adhesion molecule highly related to L1. It contains structure plan of six extracellular C2-type immunoglobulin (Ig) domains followed by five fibronectin typeⅢ domains linked by a single membrane-spanning region to a short cytoplasmic domain. The extracellular portion of CHL1 is higyly glycosylated and involved them in hemophilic disease. |
| Reference |
