Human DCXR / HCR2 Protein (His Tag)
DCR,HCR2,HCRII,KIDCR,P34H,PNTSU,SDR20C1,XR
- 100ug (NPP2076) Please inquiry
| Catalog Number | P14562-H07E |
|---|---|
| Organism Species | Human |
| Host | E. coli |
| Synonyms | DCR,HCR2,HCRII,KIDCR,P34H,PNTSU,SDR20C1,XR |
| Molecular Weight | The recombinant human DCXR consists of 259 amino acids and predicts a molecular mass of 27.8 KDa. It migrates as an approximately 29 KDa band in SDS-PAGE under reducing conditions. |
| predicted N | His |
| SDS-PAGE | 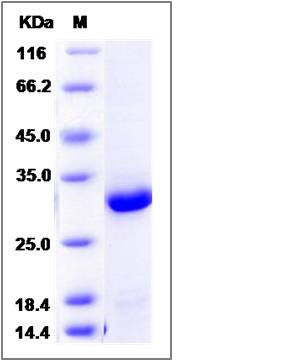 |
| Purity | > 95 % as determined by SDS-PAGE |
| Protein Construction | A DNA sequence encoding the mature form of human DCXR (Q7Z4W1) (Met1-Cys244) was expressed with a polyhistidine tag at the N-terminus. |
| Bio-activity | |
| Research Area | Cancer |Signal transduction |Metabolism |Types of disease |Metabolism in Cancer |
| Formulation | Lyophilized from sterile PBS, 20% Glycerol, pH 7.4. 1. Normally 5 % - 8 % trehalose and mannitol are added as protectants before lyophilization. Specific concentrations are included in the hardcopy of COA. |
| Background | DCXR, also known as HCR2, belongs to the short-chain dehydrogenases/reductases (SDR) family. It is highly expressed in kidney, liver and epididymis. In the epididymis, DCXR is mainly expressed in the proximal and distal sections of the corpus region. HCR2 is weakly or not expressed in brain, lung, heart, spleen and testis. DCXR catalyzes the NADPH-dependent reduction of several pentoses, tetroses, trioses, alpha-dicarbonyl compounds and L-xylulose. DCXR participates in the uronate cycle of glucose metabolism. It may play a role in the water absorption and cellular osmoregulation in the proximal renal tubules by producing xylitol, an osmolyte, thereby preventing osmolytic stress from occurring in the renal tubules. |
| Reference |
