Human OTUB2 Protein (His Tag)
C14orf137,OTB2,OTU2
- 100ug (NPP2381) Please inquiry
| Catalog Number | P13177-H07E |
|---|---|
| Organism Species | Human |
| Host | E. coli |
| Synonyms | C14orf137,OTB2,OTU2 |
| Molecular Weight | The recombinant human OTUB2 consisting of 249 amino acids and has a calculated molecular mass of 29 kDa. It migrates as an approximately 30 kDa band in SDS-PAGE under reducing conditions. |
| predicted N | Met |
| SDS-PAGE | 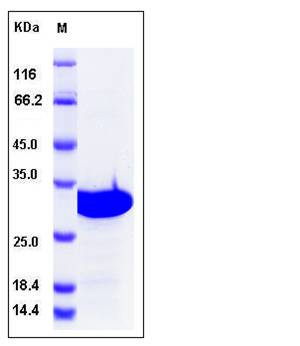 |
| Purity | > 97 % as determined by SDS-PAGE |
| Protein Construction | A DNA sequence encoding the human OTUB2 (Q96DC9-1) (Met 1-His 234) was expressed, with a polyhistide tag at the N-terminus. |
| Bio-activity | |
| Research Area | Epigenetics |Histone Modifying Enzymes |Ubiquitylation |Deubiquitination |
| Formulation | Lyophilized from sterile PBS, 10% glycerol, pH 7.5 1. Normally 5 % - 8 % trehalose and mannitol are added as protectants before lyophilization. Specific concentrations are included in the hardcopy of COA. |
| Background | Otubain 2 (OTUB2) is a member of DUBs that belong to the ovarian tumour (OTU) superfamily of proteins which consists of a five-stranded β-sheet sandwiched in between a small helical amino-terminal region consisting of α1 and α2, and a large helical region comprised of α3-α10. Like other DUBs, otubain 2 (OTUB2) cleaves proteins precisely at the ubiquitin-protein bond so that ubiquitylation process can be reversed and regulated. Otubain 2 (OTUB2)'s active-site cleft is sterically occluded by a novel loop conformation resulting in an oxyanion hole, which consists uniquely of backbone amides. Furthermore, the residues that orient and stabilize the active-site histidine of otubain 2 (OTUB2) are different from other cysteine proteases. This reorganization of the active-site topology provides a possible explanation for the low turnover and substrate specificity of the otubains. |
| Reference |
