Human RPE / RPE2-1 Protein (His Tag)
RPE2-1
- 100ug (NPP4223) Please inquiry
| Catalog Number | P11970-H08H |
|---|---|
| Organism Species | Human |
| Host | Human Cells |
| Synonyms | RPE2-1 |
| Molecular Weight | The recombinant human RPE consists of 239 amino acids and has a calculated molecular mass of 26.4 kDa as estimated in SDS-PAGE under reducing conditions. |
| predicted N | Met 1 |
| SDS-PAGE | 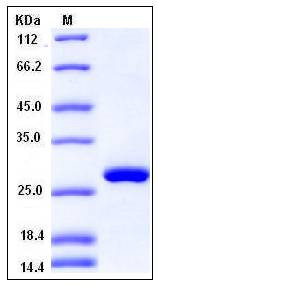 |
| Purity | > 97 % as determined by SDS-PAGE |
| Protein Construction | A DNA sequence encoding the human RPE isoform 1 (NP_954699.1) (Met 1-Arg 228) was expressed, with a polyhistidine tag at the C-terminus. |
| Bio-activity | |
| Research Area | Immunology |Signal Transduction |Metabolism |Pathways and Processes |Metabolic signaling pathways |Carbohydrate metabolism | |
| Formulation | Lyophilized from sterile PBS, pH 7.4 1. Normally 5 % - 8 % trehalose, mannitol and 0.01% Tween80 are added as protectants before lyophilization. Specific concentrations are included in the hardcopy of COA. |
| Background | The "ribulose phosphate binding" superfamily defined by the Structural Classification of Proteins (SCOP) database is considered the result of divergent evolution from a common (beta/alpha)(8)-barrel ancestor. The superfamily includes d-ribulose 5-phosphate 3-epimerase (RPE), orotidine 5'-monophosphate decarboxylase (OMPDC), and 3-keto-l-gulonate 6-phosphate decarboxylase (KGPDC). Replication of the human genome requires the activation of thousands of replicons distributed along each one of the chromosomes. Each replicon contains an initiation, or origin, site, at which DNA synthesis begins. In enzymology, a L-ribulose-5-phosphate 3-epimerase is an enzyme that catalyzes the chemical reaction L-ribulose 5-phosphate to L-xylulose 5-phosphate. Hence, RPE has one substrate, L-ribulose 5-phosphate, and one product, L-xylulose 5-phosphate. RPE belongs to the family of isomerases, specifically those racemases and epimerases acting on carbohydrates and derivatives. The systematic name of this enzyme class is L-ribulose-5-phosphate 3-epimerase. Other names in common use include L-xylulose 5-phosphate 3-epimerase, UlaE, and SgaU. |
| Reference |
