Human SIRT1 / SIR2L1 Protein (His Tag)
SIR2L1
- 100ug (NPP4273) Please inquiry
| Catalog Number | P11748-H07E |
|---|---|
| Organism Species | Human |
| Host | E. coli |
| Synonyms | SIR2L1 |
| Molecular Weight | The recombinant human SIRT1 consists of 562 amino acids and has a calculated molecular mass of 62.8 kDa. |
| predicted N | Met |
| SDS-PAGE | 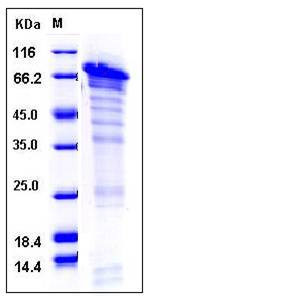 |
| Purity | > 65 % as determined by SDS-PAGE |
| Protein Construction | A DNA sequence encoding the human SIRT1 (Met 193-Ser 747) was expressed with a polyhistide tag at the N-terminus. |
| Bio-activity | |
| Research Area | Immunology |Signal Transduction |Metabolism |Types of disease |Metabolism in Obesity |
| Formulation | Lyophilized from sterile PBS, pH 7.4, 10% glycerol 1. Normally 5 % - 8 % trehalose, mannitol and 0.01% Tween80 are added as protectants before lyophilization. Specific concentrations are included in the hardcopy of COA. |
| Background | SIRT1 belongs to the sirtuin family. Members of the sirtuin family are characterized by a sirtuin core domain and grouped into four classes. SIRT1 is included in class I of the sirtuin family. It is a NAD-dependent protein deacetylase, which regulates processes such as apoptosis and muscle differentiation by deacetylating key proteins. It deacetylates 'Lys-382' of p53/TP53 and impairs its ability to induce proapoptotic program and modulate cell senescence. SIRT1 also deacetylates TAF1B and thereby represses rDNA transcription by the RNA polymerase I. It is involved in HES1- and HEY2-mediated transcriptional repression. SIRT1 inhibits skeletal muscle differentiation by deacetylating PCAF and MYOD1. It may serve as a sensor of the cytosolic ratio of NAD(+)/NADH, which is essential in skeletal muscle cell differentiation. It also deacetylates 'Lys-16' of histone H4 (in vitro). Component of the eNoSC (energy-dependent nucleolar silencing) complex, a complex that mediates silencing of rDNA in response to intracellular energy status and acts by recruiting histone-modifying enzymes. The eNoSC complex is able to sense the energy status of cell: upon glucose starvation, elevation of NAD(+)/NADP(+) ratio activates SIRT1, leading to histone H3 deacetylation followed by dimethylation of H3 at 'Lys-9' (H3K9me2) by SUV39H1 and the formation of silent chromatin in the rDNA locus. |
| Reference |
