Human SPG21 Protein (GST Tag)
ACP33,BM-019,GL010,MAST
- 100ug (NPP2509) Please inquiry
| Catalog Number | P10522-H09B |
|---|---|
| Organism Species | Human |
| Host | Baculovirus-Insect Cells |
| Synonyms | ACP33,BM-019,GL010,MAST |
| Molecular Weight | The recombinant human SPG21/GST chimera consists of 533 amino acids and predicts a molecular mass of 61 kDa which is also estimated by SDS-PAGE. |
| predicted N | Met |
| SDS-PAGE | 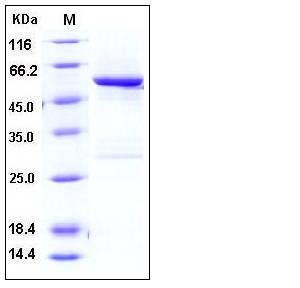 |
| Purity | > 90 % as determined by SDS-PAGE |
| Protein Construction | A DNA sequence encoding the full length of human SPG21 (NP_057714.1) (Met 1-Gln 308) was expressed with the GST tag at the N-terminus. |
| Bio-activity | |
| Research Area | Immunology |Adaptive Immunity |T Cell |Non-CD of T cell |
| Formulation | Lyophilized from sterile 50mM Tris, 100mM NaCl, pH 8.0, 10% glycerol 1. Normally 5 % - 8 % trehalose, mannitol and 0.01% Tween80 are added as protectants before lyophilization. Specific concentrations are included in the hardcopy of COA. |
| Background | Spastic paraplegia 21 (SPG21), also known as acid Cluster Protein 33 (ACP33) and Mast syndrome protein, is a member of the AB hydrolase superfamily. Human SPG21 is a 308 amino acid residue protein widely expressed in all tissues, including heart, brain, placenta, lung, liver, skeletal muscle, kidney and pancreas. SPG21 binds to the hydrophobic C-terminal amino acids of CD4 which are involved in repression of T cell activation via the noncatalytic alpha/beta hydrolase fold domain. SPG21 thus is proposed to play a role as a negative regulatory factor in CD4-dependent T-cell activation of CD4. Defects in SPG21 are the cause of spastic paraplegia autosomal recessive type 21, also known as Mast syndrome, a neurodegenerative disorder characterized by a slow, gradual, progressive weakness and spasticity of the lower limbs. Rate of progression and the severity of symptoms are quite variable. SPG21 is also associated with dementia and other central nervous system abnormalities. |
| Reference |
