Mouse Smad2 Protein (His & GST Tag)
7120426M23Rik,Madh2,Madr2,mMad2,Smad-2
- 100ug (NPP1681) Please inquiry
| Catalog Number | P50727-M20B |
|---|---|
| Organism Species | Mouse |
| Host | Baculovirus-Insect Cells |
| Synonyms | 7120426M23Rik,Madh2,Madr2,mMad2,Smad-2 |
| Molecular Weight | The recombinant mouse SMAD2/GST chimera consists of 703 amino acids and has a calculated molecular mass of 80 kDa. The recombinant protein migrates as an approximately 90 kDa band in SDS-PAGE under reducing conditions. |
| predicted N | Met |
| SDS-PAGE | 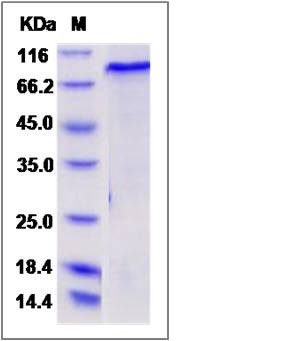 |
| Purity | > 90 % as determined by SDS-PAGE |
| Protein Construction | A DNA sequence encoding the mouse SMAD2 (Q62432-Isoform Long) (Ser2-Ser467) was expressed with the N-terminal polyhistidine-tagged GST tag at the N-terminus. |
| Bio-activity | |
| Research Area | Signaling |Signal Transduction |Transcription Factors and Regulators |Smad Transcription Factors |
| Formulation | Lyophilized from sterile 20mM Tris, 500mM Nacl, pH 8.0, 10% glycerol, 3mM DTT 1. Normally 5 % - 8 % trehalose and mannitol are added as protectants before lyophilization. Specific concentrations are included in the hardcopy of COA. |
| Background | SMAD2 is a member of the SMAD family. Members of this family mediate signal transduction by the TGF-beta/activin/BMP-2/4 cytokine superfamily from receptor Ser/Thr protein kinases at the cell surface to the nucleus. SMAD2 mediates the signal of the TGF-beta, and therefore regulates multiple cellular processes, such as cell proliferation, apoptosis, and differentiation. SMAD2 is recruited to the TGF-beta receptors through its interaction with the SMAD anchor for receptor activation (SARA) protein. SMAD2 is the downstream signal transducers of TGF-beta-1 in human dental pulp cells. In response to TGF-beta signal, this protein is phosphorylated by the TGF-beta receptors. Phosphorylated SMAD2 is able to form a complex with SMAD4 or SARA. These complexes accumulate in the cell nucleus, where they are directly participating in the regulation of gene expression. |
| Reference |
