Rat CLP1 / COLEC12 Protein (His Tag)
Clp1, Nsr2, COLEC12
- 100ug (NPP2889) Please inquiry
| Catalog Number | P80394-R07B |
|---|---|
| Organism Species | Rat |
| Host | Baculovirus-Insect Cells |
| Synonyms | Clp1, Nsr2, COLEC12 |
| Molecular Weight | The recombinant ratCOLEC12 consists of 658 amino acids and predicts a molecular mass of 72.4 KDa. It migrates as an approximately 90 KDa band in SDS-PAGE under reducing conditions. |
| predicted N | His |
| SDS-PAGE | 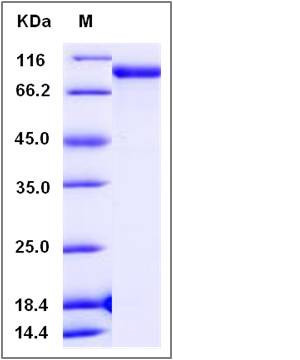 |
| Purity | > 99 % as determined by SDS-PAGE |
| Protein Construction | A DNA sequence encoding the mature form of rat COLEC12 (Q4V885) (Ala101-Leu742) was expressed,with a polyhistide tag at the N-terminus. |
| Bio-activity | |
| Research Area | Cancer |Invasion microenvironment |Adhesion molecule |Cell adhesion |Lectin |C-tyep lectin | |
| Formulation | Lyophilized from sterile 20mM Tris, 500mM NaCl, pH 8.0 1. Normally 5 % - 8 % trehalose, mannitol and 0.01% Tween80 are added as protectants before lyophilization. Specific concentrations are included in the hardcopy of COA. |
| Background | CLP1, also known as COLEC12, is a scavenger receptor that displays several functions associated with host defense. It contains 1 C-type lectin domain and 3 collagen-like domains. CLP1 is strongly expressed in placenta and moderately expressed in heart, skeletal muscle, small intestine and lung. It promotes binding and phagocytosis of Gram-positive, Gram-negative bacteria and yeast. CLP1 mediates the recognition, internalization and degradation of oxidatively modified low density lipoprotein (oxLDL) by vascular endothelial cells. It binds to several carbohydrates including Gal-type ligands, D-galactose, L- and D-fucose, GalNAc, T and Tn antigens in a calcium-dependent manner and internalizes specifically GalNAc in nurse-like cells. It binds also to sialyl Lewis X or a trisaccharide and asialo-orosomucoid (ASOR). CLP1 may also play a role in the clearance of amyloid beta in Alzheimer disease. |
| Reference |
|
